High-Throughput SARS-CoV-2 Detection by NGS
SARS-CoV-2 Genomic Surveillance
The FGCZ also supports the currently largest SARS-CoV-2 sequencing effort in Switzerland as part of the Swiss Viollier Sequencing Consortium, led by ETH’s D-BSSE Computational Evolution group.
For more information please visit these sites:
Swiss Viollier Sequencing Consortium
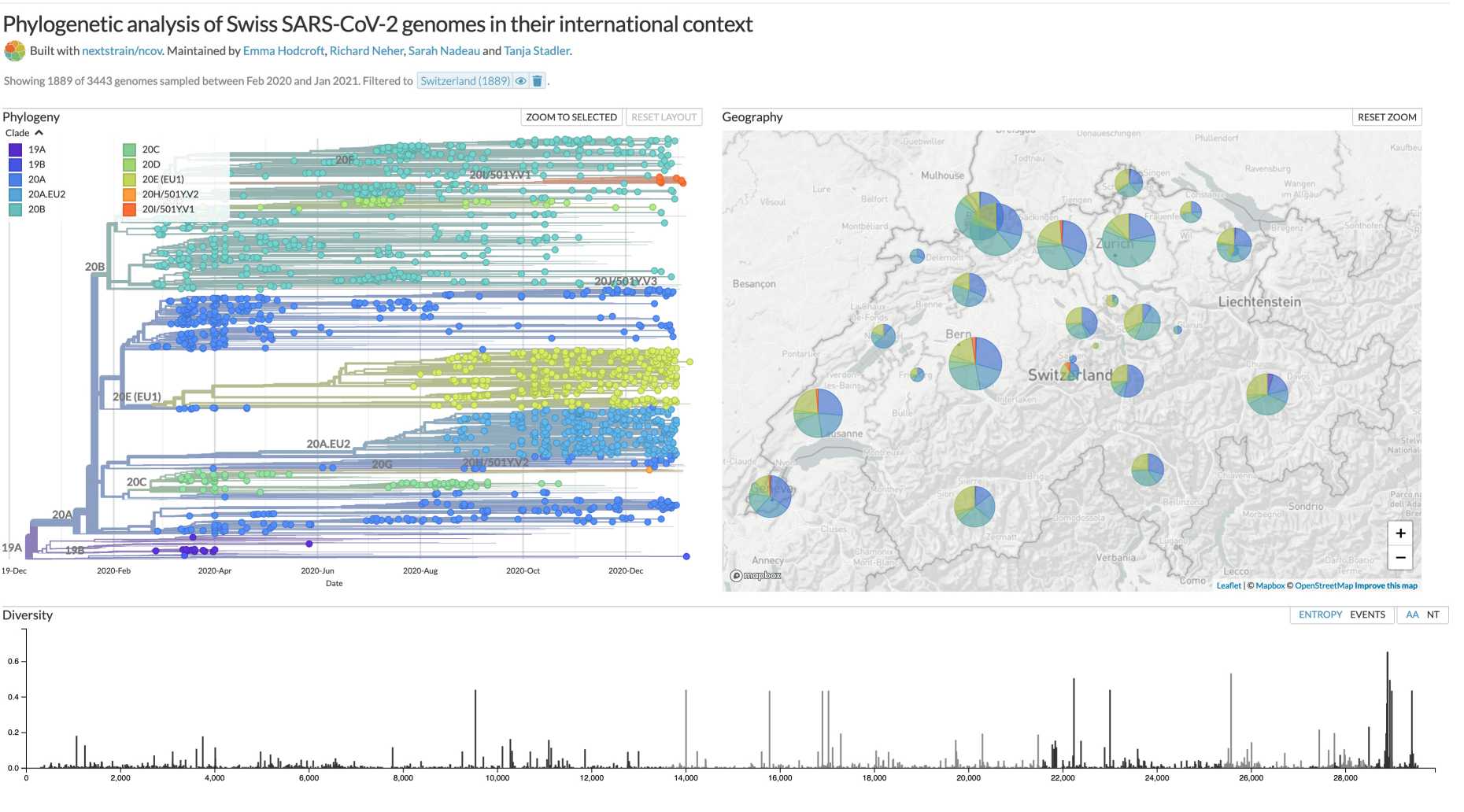
external page Phylogenetic analysis of Swiss SARS-CoV-2 genomes in their international context
external page Swiss National Covid-19 Science Task Force

The FGCZ has developed an initial protocol for next generation sequencing-based coronavirus detection , called HiDRA-seq. It is a rapidly implementable, high throughput, and scalable solution that uses NGS lab infrastructure and reagents for population-scale SARS-CoV-2 testing. This method is based on the use of indexed oligo-dT primers to generate barcoded cDNA from a large number of patient samples. From this, highly multiplexed NGS libraries are prepared targeting SARS-CoV-2 specific regions and sequenced. The low amount of sequencing data required for diagnosis allows the combination of thousands of samples in a sequencing run, while reducing the cost to approximately 2 CHF/EUR/USD per RNA sample. In our preprint in BioRxiv that can be read external page here, we describe in detail the first version of the protocol, which can be further improved in the future to increase its sensitivity and to identify other respiratory viruses or analyze individual genetic features associated with disease progression.