News
June 2025

Second Edition of Student-on-Stage
Last year, we hosted the first-ever FGCZ Students-on-Stage event, which received enthusiastic feedback from both students and staff.
It provided a valuable platform for students to present their omics-related projects and gave us the opportunity to learn from a broad spectrum of biological research, inspiring future developments.
This year, we proudly held another successful edition. A heartfelt thank you to all the participants, the organizer, the jury, and especially the brilliant students.
We are pleased to see how FGCZ can support such a wide range of biological research through student presentations, while also remaining humble in the face of the many unknowns that biology continues to reveal.
All the presentations were of such high quality that the jury faced a difficult task in selecting a winner. In the end, the award for best presentation went to bumblebee expert Priska Flury.
We would also like to extend our sincere thanks to all the outstanding presenters: Emma Alessandri, Andrea Roggo, Laura Much, Weijia (Natalie) Zhong, Vladyslav Korobeynyk, Stefanie Schürch, Marta Mazurkiewicz, Maria Andreea Dimitriu, Cyrill Hofer, Debleena Basu, Guillermo de la Vega Barranco, Houria Ech-cherif, Jana Bangerter, and Josephine von Kempis. Your presentations were truly impressive — great job!
We hope everyone enjoyed the event, and we’re already looking forward to welcoming you back next year for the third edition!

April 2025

We are excited to announce a new strategic collaboration between the Metabolomics and Bioinformatics Units at the Functional Genomics Center Zurich (FGCZ) and mzio GmbH, formalized through a support agreement under the mzmine PRO license.
At the FGCZ, the Metabolomics Unit brings a wealth of experience and expertise in providing state-of-the-art user-centered analytical technologies, from supporting robust experimental design to implementing advanced LC-MS approaches that generate high-quality data.
Our Bioinformatics Unit complements this by supporting computational metabolomics needs from QC dashboards, LC-MS data analysis, visualization, and report generation apps integrated in our data management system, external page B-Fabric.
Our combined mission is to support biological and biomedical research through reliable, reproducible metabolomics and lipidomics analysis, fulfilling our goals of impact, efficiency, and innovation.
Through this collaboration with mzio, mzmine — an open-source platform with over 5,500 citations with the latest publication in external page Nature Biotechnology (Schmid, Heuckeroth, Korf et al., 2023) — will be supported and integrated into FGCZ’s metabolomics services in the following months. Its implementation and further optimization will be supported via the mzmine PRO license. Through this partnership, we aim to:
- Provide access to leading open-source tools for untargeted metabolomics and lipidomics
- Ensure transparency in every step of the data analysis pipeline, aligned with FAIR principles
- Streamline data processing and analysis within FGCZ infrastructure
- Empower users with intuitive, interactive tools for exploring their data
- Enhance reproducibility and collaboration via full integration into B-Fabric
This partnership is a step forward in our journey to democratize metabolomics and empower the research community in Zurich and beyond with the best in the field.
Stay tuned for updates on our user-centered innovations!
March 2025

Take Two!
After last year's successful 'Students on Stage', where 12 talented students shared their research with us, we are now entering the second round.
We look forward to exploring many more projects enriched with OMIC data from the FGCZ this year.
Would you like to be part of it?
As part of a mini-symposium on 11 June, Master and PhD students from UZH & ETH can present their exciting projects in an oral presentation and have the chance to win a great prize.
But of course, what is a conference without an audience?
Anyone interested in discovering the diverse and exciting projects of our local research community is warmly welcome. Space is limited, so external page reserve your seat today.
external page Abstract submission is open until 16 May!
January 2025

We are very proud of our latest Proteomics publication external page A Multiyear Longitudinal Harmonization Study of Quality Controls in Mass Spectrometry Proteomics Core Facilities!
In this study, we performed a coordinated effort to establish a common Quality Control framework within the proteomics core facilities of the Core4Life alliance, with the goal to enhance the consistency of quality control results.
Over the course of 4 years, we:
- Developed a New Common Quality Control Standard
- Acquired over 9000 raw files on 30 LC-MS systems
- Used QCloud as a Common Quality Control Processing Software
- Exchanged information on LC-MS-related problems and reduce the variability in our LC-MS systems
- Defined Reference Values to share with the community
June 2024

The FGCZ has been participating in the Green Labs pilot program of external page Leaf (Laboratory Efficiency Assessment Framework) and was awarded a Gold Certificate for our achievements in Lab Sustainability.
This is an onging pilot project in which both the ETH Zurich and the external page University of Zurich are partaking.
Maybe your lab should join too?
April 2024

On June 13th, 2024 the Functional Genomics Center Zurich will host the first edition of the "Students on Stage" event. We look forward to welcoming many ETH/UZH students to present their OMICS-based research to the local community. The event will take place at our Center in a conference-like format. Master and PhD students who have conducted OMICS research in collaboration with the FGCZ (either in user lab or service mode) are eligible to present their work and are warmly invited to submit an abstract using this external page link. Everyone is welcome, from the experienced PhD student in their final year to the newcomer. Take advantage of this opportunity to make your research visible! Best part, a prize will be awarded for the best oral presentation.
Abstract due by May 16th, 2024.
Date:
Thursday June 13th, 2024, 9:00 – 12:00
Location:
Functional Genomics Centre (FGCZ)
Winterthurerstrasse 190 CH-8057 Zürich, Seminar Room Y59, Floor G
If you have any questions, please contact
August 2023

Come join us at Scientifica 2023, the largest science festival in Switzerland organized by ETH and UZH!
How do organisms work? In the fascinating interplay of life, the interactions between DNA, RNA, proteins and metabolites direct the complex processes that control the functions of living organisms.
The FGCZ is happy to welcome you on Saturday 2nd and Sunday the 3rd of September for tours of our center in German (12:00-13:00) and English (14:00-15:00). The event is for families, so kids are more than welcome and will have the chance to "work" in our labs!
external page Registration is required on the same day of the event, and places are limited!
August 2023

UZH Life Sciences Core Facility Day
“Unleashing Synergy: Connecting Life Sciences Technologies”
Join us on Tuesday 22 August 2023, the UZH Life Sciences Core Facilities Day will take place from 14:00 to 16:00 in the Lichthof at Irchel.
May 2023

New Publication:
Plastic-eating fungi discovered in Swiss Alps
Bacteria and fungi have recently been discovered in the Swiss Alps showing their ability to digest biodegradable plastics, even at low temperatures. Two studies have revealed the impact of plastics on the alpine soil microbiome, unveiling numerous novel microbial taxa with the capacity to break down biodegradable plastic films, including dispersed PUR and PBAT. These findings lay a robust foundation to emphasize the role of biodegradable polymers in a circular plastic economy.
FGCZ genomics expert, Dr. Weihong Qi, employed a shotgun metagenomics method to verify the effects of polyethylene, as well as the biodegradable films Ecovio and BI-OPL, which were buried in alpine soils for 5 months, on the genetic potential of the soil microbiome.
The paper can be read here:
Journal of Hazardous Materials 441 (2023) 129941
Media reports on
external page Swissinfo and external page BBC news
April 2023

FGCZ Seminar series
Tuesday April 25th, 2023, 16:00-17:00
Towards a tissue specific and structurally resolved human interactome
Prof. Dr. Pedro Beltrao
Department of Biology, ETH Zurich
Download More information and how to register (PDF, 182 KB)
April 2023

the FGCZ on Social Media
We are pleased to announced the launch of our external page FGCZ LinkedIn page and external page FGCZ Twitter account.
Through these accounts, we will share with you our news, updates about publications, technologies, teaching and outreach activities. We will also introduce you to our portfolio, and most importantly to our staff; the team behind the scenes and without whom none of our work would be possible.
We look forward to your positive engagement and are always happy to support you with multiomics in your projects (for any project-related communications, please use the standard external page B-Fabric portal using your project number, or the email addresses on our website for general enquiries).
April 2023

NanoString Seminar
On Wednesday 26th April 2023 NanoString will stop by the FGCZ as part of their 7 Country external page Europe Roadshow to discuss how to elevate your single-cell biology research by measuring and visualizing the full RNA reactome in situ. Plus, don’t miss an exclusive demo of AtoMx™ Spatial Informatics Platform (SIP), the only cloud-based, fully-integrated informatics platform for spatial biology.
Click Download here for the full program and how to register (PDF, 311 KB).
March 2023

Element Biosciences Seminar
We are very happy to welcome Molly He, Element Biosciences CEO &
Co-Founder for a seminar where she will introduce the AVITI system
Element Biosciences has reinvented DNA sequencing with the introduction
of the AVITI system. Using a fundamentally different DNA sequencing
method, the AVITI system flips the script with a benchtop instrument
that provides higher-quality data at a much lower cost. Learn about
avidity sequencing, common use cases for WGS, RNA sequencing, and
targeted sequencing applications.
Date:
March 30, 2023
Location:
Functional Genomics Center
ETH Zurich/University of Zurich
Winterthurer Strasse 190 / Y59 G Floor
10:00 – 10:15
Ewoud Ouwerkerk, General Manager EMEA, Element Biosciences
Introduction to Element Biosciences
10:15-11:00
Molly He, CEO of Element Biosciences
“Avidity Sequencing on the AVITI System - Sequencing Reimagined”
11:00 – 11:20
Q&A
11:20 – 12:00
Come together with Coffee & Snacks
March 2023

The FGCZ is featured in a new Universty of Zurich article focused on teaching
UZH Teaching Fund – Part 1: Research-based teaching
external page Taking the Plunge into Research
February 2023

LS2 Annual Meeting 2023 - "Single Cell Proteomics: are we there yet?"
Friday 17 February 2023 / 14:05 – 15:50, Irchel Campus Lecture Hall G40
Chairs: Dr. Paolo Nanni (FGCZ, ETH/UZH) and Dr. Maria Pavlou (EPFL)
Are you attending the external page LS2 Annual Meeting 2023 “Life on Earth: Coping with Challenges”? If yes, do not miss the Proteomics Symposium. The program includes keynote speakers, an industry talk, selected speakers from abstracts and a final Round Table Discussion.
Program:
- Dr. Edward Emmott (Centre for Proteome Research, University of Liverpool, UK): "Prioritised SCoPE2-based analysis of post-translational modifications at single-cell level"
- Dr. Torsten Müller (Bruker Switzerland AG): "On the benefits and challenges of single cell proteomics"
- Prof. Bernd Bodenmiller (University of Zurich): "Highly multiplexed imaging of tissues with subcellular resolution by imaging mass cytometry"
- Daniel Gonzalez-Bohorquez (University of Zurich) "Low input proteomics to understand cell division history and metabolic disruptions in human forebrain organoids”
- Round Table Discussion (30 min)
January 2023

EuBIC-MS Developers Meeting 2023
15-20 January 2023
Co-organized with the FGCZ, the European Bioinformatics Community for Mass Spectrometry (EuBIC-MS) is having a external page Developers Meeting 2023. It takes place in the ETH meeting platform Congressi Stefano Franscini (CSF), Monte Verità, Ticino, Switzerland.
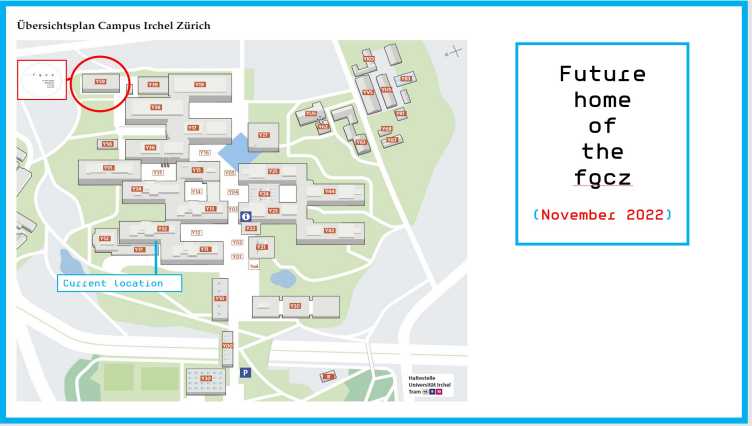
November 2022

FGCZ Open Lab
Friday, November 25, from 2 to 5 pm
The FGCZ opens its new labs and offices in building Y59 on the Irchel Campus of UZH.
On Friday, November 25, from 2 to 5 pm our new facility is open to interested users and researchers who want to know more about our technologies and research support options.
Expert analytical and bioinformatics staff will be available throughout the center and inform about our offerings to support multi-omics research projects.
Topics and technologies include
- Genomics (sequencing technologies, single cell and spatial transcriptomics, genome engineering, and more)
- Proteomics (proteome and PTM profiling, interaction and spatial proteomics, biomolecule characterization, and more)
- Metabolomics (untargeted and targeted metabolomics and lipidomics, spatial applications, and more)
- Biophysics (kinetic interaction analysis, microarray studies, protein quality determination, and more)
To register, please contact us at
July 2022

10x Genomics and Functional Genomics Center Zürich (FGCZ) are Sponsoring a Grant Program for Innovative Investigators at ETH Zürich and University of Zürich
We are pleased to announce the external page 10x Genomics Visium Spatial Gene Expression Grant Program. This award covers library preparation and bioinformatic support for a pilot project using Visium Spatial for FFPE Gene Expression from 10x Genomics.
May 2022

Epic-XS Update
As a member of the external page EPIC-XS network, the proteomics team at the FGCZ has almost reached its full capacity of pledged access days. More than 40 projects have been accepted since June 2019 and over 310 days of sample processing, data acquisition and data analysis were provided for collaborators across Europe until now. Projects so far have spanned multiple scientific areas covering agricultural and plant science, as well as basic and biomedical research. For the remainder of access days available, we will focus on projects submitted through the joint call with external page EASI-Genomics.
April 2022

Generating a reference-quality assembly for the highly complex Cassava genome
As reported in external page GigaScience, the sequencing team at the Functional Genomics Center Zurich, in collaboration with the Gruissem laboratory at the Institute of Molecular Plant Biology in ETH Zurich, has applied cutting-edge long-range sequencing techniques and data analysis expertise to generate the most accurate Cassava genome assembly yet. The reference-quality, haplotype-resolved genome assembly also enabled the construction of cassava pan-genome, which accelerates breeding of this important crop. A more detailed report is also featured in the ETHZ D-BIOL News Channel.
February 2022

Our review “external page Mass Spectrometry in Proteomics: Technologies, Methods, and Research Applications for the Life Sciences” (Chimia 76 (2022) 73–80) is now online!
In this review we discussed the importance of mass spectrometry for the life science sector with a special focus on the most relevant current applications in the field of proteomics. Moreover, we commented on the factors that research groups should consider when setting up a mass spectrometry laboratory, and on the fundamental role played by academic core facilities and industrial service providers.
The manuscript is part of the Special Chimia issue Vol. 76 No. 1-2 (2022): “Mass Spectrometry at Swiss Academic and Industrial Institutions”.
January 2022

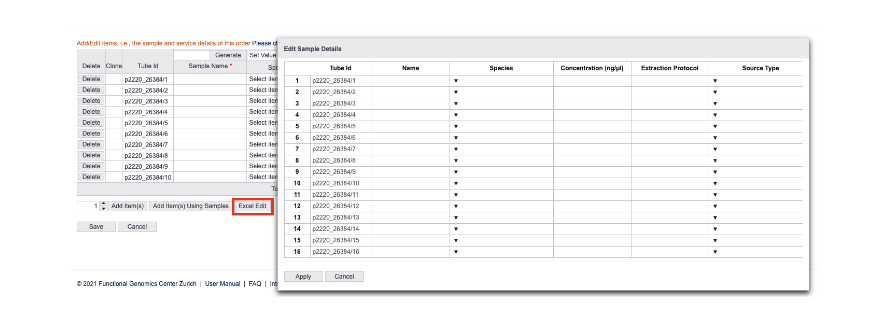
The number of samples processed at the FGCZ is constantly increasing, so we have now made creating orders much easier in external page B-Fabric with the following new functionalities:
- Plate submission option (only for NGS services): When you have more than 30 samples, they can now be submitted in 96-well plates for easy handling. The detailed instructions can be found external page here.
- Excel edit option (all orders): You can now enter information about your samples (Sample Name, Species, Concentration, etc) by copy-pasting from Excel.
December 2021

The scientific board of the Congressi Stefano Franscini accepted the FGCZ application for the external page EuBIC-MS Developers’ Meeting on
January 15th-20th, 2023
(information on the external page previous event, 2020)
November 2021

Major Milestone!
The FGCZ has surpased 1000 runs on the Novaseq.
We are sequencing everything from viruses to mummies!
*Congratulations to the Genomics team*
July 2021

Thursday, July 15th, 15:00 - 16:30
FGCZ will be presenting in the third Webinar of the LS2 series “external page A Proteomics Tour of Switzerland”
The Zurich Proteomics Hub - Deeper insights with multi-dimensional prototyping
Organized by the LS2 Proteomics Section, these virtual sessions are bringing us to different cities in Switzerland to discover what Proteomics is all about there.
June 2021
New Publication
Embo Reports
external page Quality standards in proteomics research facilities
March 2021

New Publication
Journal of Proteome Research
external page The rawrr R Package: Direct Access to Orbitrap Data and Beyond
March 2021
Due to the FGCZ's continued effort in supporting SARS-CoV-2 genomic surveillance sequencing, capacities for the processing of Illlumina-based NGS orders are currently limited, which can lead to delays in the delivery of research project data – please check with your order or project coach for details.
December 2020
FGCZ Genomics Platform performs large-scale SARS-CoV-2 genome sequencing to identify virus variants
The FGCZ Genomics Platform uses Illumina high throughput sequencing technology to sequence the genomes of multiple hundred human RNA samples per week from positively tested individuals. In collaboration with D-BSSE, recently emerged external page new variants have been positively identified
The FGCZ continues to use and establish NGS-based analysis methods for pandemic control, in parallel to supporting all its user projects with the technology.
November 2020

New Publication
Gas-Phase Fragmentation of ADP-Ribosylated Peptides: Arginine-Specific Side-Chain Losses and Their Implication in Database Searches
ADP-ribosylation is a reversible post-translational modification of proteins that has been linked to many biological processes. The identification of ADP-ribosylated proteins and particularly of their acceptor amino acids remains a major challenge. The attachment sites of the modification are difficult to localize by mass spectrometry (MS) because of the labile nature of the linkage and the complex fragmentation pattern of the ADP-ribose in MS/MS experiments. In this study we performed a comprehensive analysis of higher-energy collisional dissociation (HCD) spectra acquired from ADP-ribosylated peptides which were modified on arginine, serine, glutamic acid, aspartic acid, tyrosine, or lysine residues. In addition to the fragmentation of the peptide backbone, various cleavages of the ADP-ribosylated amino acid side chains were investigated. We focused on gas-phase fragmentations that were specific either to ADP-ribosylated arginine or to ADP-ribosylated serine and other O-linked ADP-ribosylations. The O-glycosidic linkage between ADP-ribose and serine, glutamic acid, or aspartic acid was the major cleavage site, making localization of these modification sites difficult. In contrast, the bond between ADP-ribose and arginine was relatively stable. The main cleavage site was the inner bond of the guanidine group, which resulted in the formation of ADP-ribosylated carbodiimide and of ornithine in place of modified arginine. Taking peptide fragment ions resulting from this specific cleavage into account, a considerably larger number of peptides containing ADP-ribosylated arginine were identified in database searches. Furthermore, the presence of diagnostic ions and of losses of fragments from peptide ions allowed us, in most cases, to distinguish between ADP-ribosylated arginine and serine residues.
external page DOI:10.1021/jasms.0c00040
September 2020
CURRENT FGCZ OPERATIONS
The FGCZ is fully operational in all sections. Special precautions have been put in place in line with the directives of the University of Zurich and ETH Zurich.
All services are running with close to normal capacity, limitations in terms of numbers of staff and users allowed to work in labs and offices are still in place. All staff, users and visitors of the FGCZ have to wear facemasks in labs and meeting rooms. User lab operations and hence access to instruments at the FGCZ is possible, in compliance with the measures of ETH, UZH and the FOPH. Autonomous and experienced users are granted access, based on capacity and exclusively after scheduling and agreement by FGCZ staff. Training and teaching activities of the FGCZ take place with a reduced on-site presence and additional virtual modules. On-site courses follow the guidelines for practicals of ETH and UZH. All bioinformatics and data analysis services, as well as user access to data and data analysis tools have been and remain fully operational. The on-site bioinformatics user lab remains closed. Bioinformatics staff is accessible via all remote communication tools of ETH and UZH.
To obtain latest information on available services and for instructions on how to submit samples under the current regime, please get in contact with your project and order coaches prior to sending materials or planning time critical experiments. Access and service information is also constantly updated at external page fgcz.ch
June 2020

High-Throughput SARS-CoV-2 Detection by NGS
The FGCZ has developed an initial protocol for next generation sequencing-based coronavirus detection , called HiDRA-seq. It is a rapidly implementable, high throughput, and scalable solution that uses NGS lab infrastructure and reagents for population-scale SARS-CoV-2 testing. This method is based on the use of indexed oligo-dT primers to generate barcoded cDNA from a large number of patient samples. From this, highly multiplexed NGS libraries are prepared targeting SARS-CoV-2 specific regions and sequenced. The low amount of sequencing data required for diagnosis allows the combination of thousands of samples in a sequencing run, while reducing the cost to approximately 2 CHF/EUR/USD per RNA sample. In our preprint in BioRxiv that can be read external page here, we describe in detail the first version of the protocol, which can be further improved in the future to increase its sensitivity and to identify other respiratory viruses or analyze individual genetic features associated with disease progression.